A systematic review of real-world applications of genome sequencing for newborn screening
Abstract
Aim: With the costs of genomic sequencing falling quickly and an ever-increasing number of clinical laboratories equipped with new-generation sequencing machines, healthcare systems around the world are getting ready to enter the era of genomic newborn screening (NBS). However, the adoption of Genomic Sequencing (GS), encompassing whole-exome sequencing (WES) and whole-genome sequencing (WGS), in NBS programs raises a number of clinical, ethical, and legal questions as well as organizational and economic challenges. This systematic review is part of a feasibility study to assess the introduction of WGS for NBS in Lombardy region with the specific aim of gathering evidence from existing pilots in the field whose results have been published.
Methods: Three different sources were identified for the selection of articles in order to obtain a various and unbiased set of publications. 33 articles were retained for analysis to answer the following questions:
1. Clinical: Does genomic sequencing demonstrate clinical utility in the context of NBS? What are the limitations of these kind of programs?
2. Societal: What are the social, ethical and psychological implications of using GS for NBS?
3. Governance: What are the legal, economic, and organizational challenges for GS-based NBS programs?
Results: There is a general consensus in the literature on the key principles that should guide the adoption of GS in NBS, such as the inclusion of actionable genes only, the need for informed consent from the parents, the right of the newborn to an open future, which means the exclusion of late-onset diseases even when those are considered treatable. However, there are still several differences in how these principles are detailed and applied.
Conclusion: Real-world evidence from a handful of pilot projects (namely BabySeq and NC-Nexus, both carried out in the USA) have been published recently; however, this evidence is not yet sufficient to put an end to the broad and animated debate on the use of GS for NBS. Ethical, legal, and social issues still constitute great challenges and major barriers to wide and uniform adoption of GS in NBS. On the clinical side, a number of issues remain unaddressed, such as the benefits and limitations of the different approaches (targeted sequencing, GS only versus GS+standard NBS), the genes/diseases to include and the frequency of incidental findings, identification of carrier status, and variants of uncertain significance (VUS). Further pilots and consultations with involved stakeholders will be necessary before GS-based NBS can be accepted and systematically implemented in national healthcare programs.
Keywords
INTRODUCTION
Newborn screening (NBS) programs have been running successfully for more than 50 years since its introduction in the 1960s. In many countries, the first disorder included in screening programs was Phenylketonuria (PKU). With the advent of Tandem Mass Spectrometry (MS/MS), the number of conditions screened increased to around 50, although with great disparities among countries[1]. The introduction of MS/MS was therefore a key driver for the expansion of the number of conditions screened, with an increase in the order of 10 folds. Now, with the costs of genomic sequencing falling quickly and an ever-increasing capacity of laboratories as more and more are getting equipped with new generation sequencing instruments, a further scale-up of NBS programs is technically possible, also in the order of 10 folds (from 50 to 500 conditions)[2-6]. However, it is important to consider that one disease can be linked to one or more genes, and for each gene, there could be several variants, pathogenic or not. A major limitation of the GS approach is that several variants cannot be classified either as non-pathogenic or pathogenic and are actually classified as variants of uncertain/unknown significance (VUS). The specific criteria for selecting the genes and the conditions to be screened are not yet unanimously accepted, even if there is a general agreement that only pathogenic or likely-pathogenic variants should be reported and the principles set by Wilson and Jungner are still basically valid[7]. Moreover, the adoption of Genomic Sequencing (GS), meaning whole-exome sequencing (WES) or whole-genome sequencing (WGS), poses a number of clinical, ethical, and legal questions[8-12] together with organizational and economic challenges[3,13-14].
This systematic review is part of a feasibility study assessing the introduction of GS for NBS in Lombardy region (Italy) and is co-funded by the regional government (Regione Lombardia) and Fondazione Telethon. The study is conducted according to the Responsible Research and Innovation (RRI) principles[15-18] and is inspired by the EUNetHTA Core Model®[19,20]. RRI principles include, among others, engagement of all societal actors, gender balance both within the research teams and in the group of consulted stakeholders, ethics, and governance, with the intent to enable a positive impact of the research on society.
Considering the nine domains of EUNetHTA Core Model®, the purpose of this review is to inform the activities of the feasibility study in the following domains while addressing relevant and associated issues:
(1) Health Problem and Current Use of the Technology with a special focus on pilot projects that tested GS for NBS;
(2) Description and technical characteristics of the technology with a focus on the discussion within the scientific community on the list of genes that should (or should not) be included in the analysis;
(3) Safety with a focus on incidental findings, false negatives, and false positives;
(4) Clinical Effectiveness trying to answer the question: What is the number of newborns per year we could expect to identify as positive?
(5) Costs and economic evaluation to investigate which methods and models were used to estimate the costs of GS-based NBS by ongoing initiatives;
(6) Ethical analysis considering in particular that in the case of NBS the patient cannot make any decision by himself/herself, as all decisions are taken by the parents;
(7) Organizational aspects - again looking at recent pilots, trying to identify the major obstacle(s) to the full deployment as part of the standard of care of a GS-NBS program;
(8) Patients and Social aspects with a focus on the acceptability of GS-based screening programs by citizens and the methodology adopted by other pilot programs to consult and engage citizens;
(9) Legal aspects to first answer the question of whether a genomic screening program could be made mandatory (as it is now for the traditional Italian NBS program) or should be voluntary.
Trying to cover all the above-mentioned issues, we selected a wide search algorithm without limiting our review to a specific domain but limiting it to newborn/neonatal screening AND WGS (that includes, as a MeSH term, WES). For results and conclusions, we grouped the above-listed domains into three main areas: Clinical (covering issues 1 to 4), Societal (covering issues 6 and 8), and Governance (covering issues 5, 7, and 9).
METHODS
Search strategy
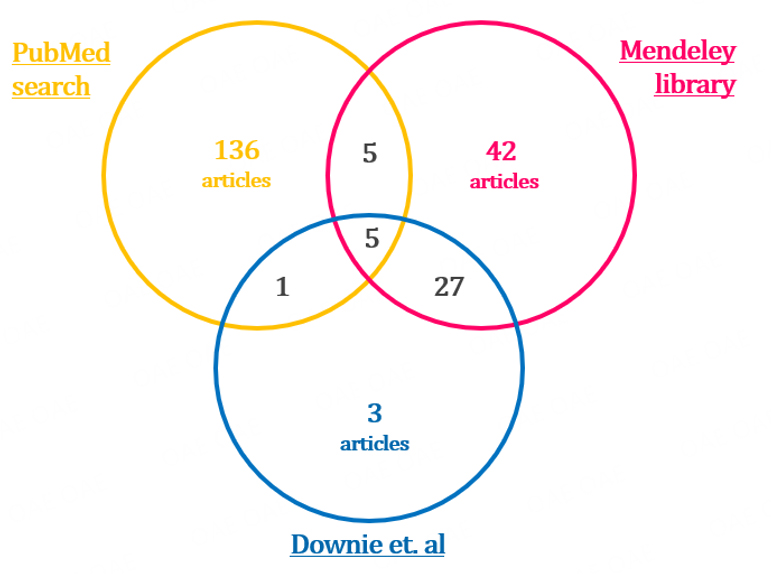
Three different sources were identified for the selection of the papers in order to obtain a various and unbiased set of articles. The sources included (1) the PubMed online database via a query performed on September 28th, 2022; (2) the Mendeley library shared within the clinicians working group; and (3) the final selection of articles that were selected for Downie et al.’s 2021 systematic review “Principles of Genomic Newborn Screening Programs: a systematic review[21]”.
The search algorithm used in PubMed was defined according to the objective of the review, i.e., to provide the practical and theoretical background for the application of WGS or WES techniques to population-wide NBS programs. The search was performed for all study types published in English, with the full texts available using MeSH terms (whole-genome sequencing) AND (neonatal screening). These MeSH terms were selected because they include all the possible synonyms, and in the case of WGS, it includes WES as well. The query on the PubMed online database with this algorithm gave 147 articles as a result.
The Mendeley library has been populated by the multidisciplinary team working on the feasibility study mentioned in the introduction. 79 articles were identified and used to guide the conception, design, and start-up phases of the study.
Downie et al.’s systematic review “Principles of Genomic Newborn Screening Programs: a systematic review” published in 2021 was considered the benchmark and the 36 final articles were included in our initial database[21].
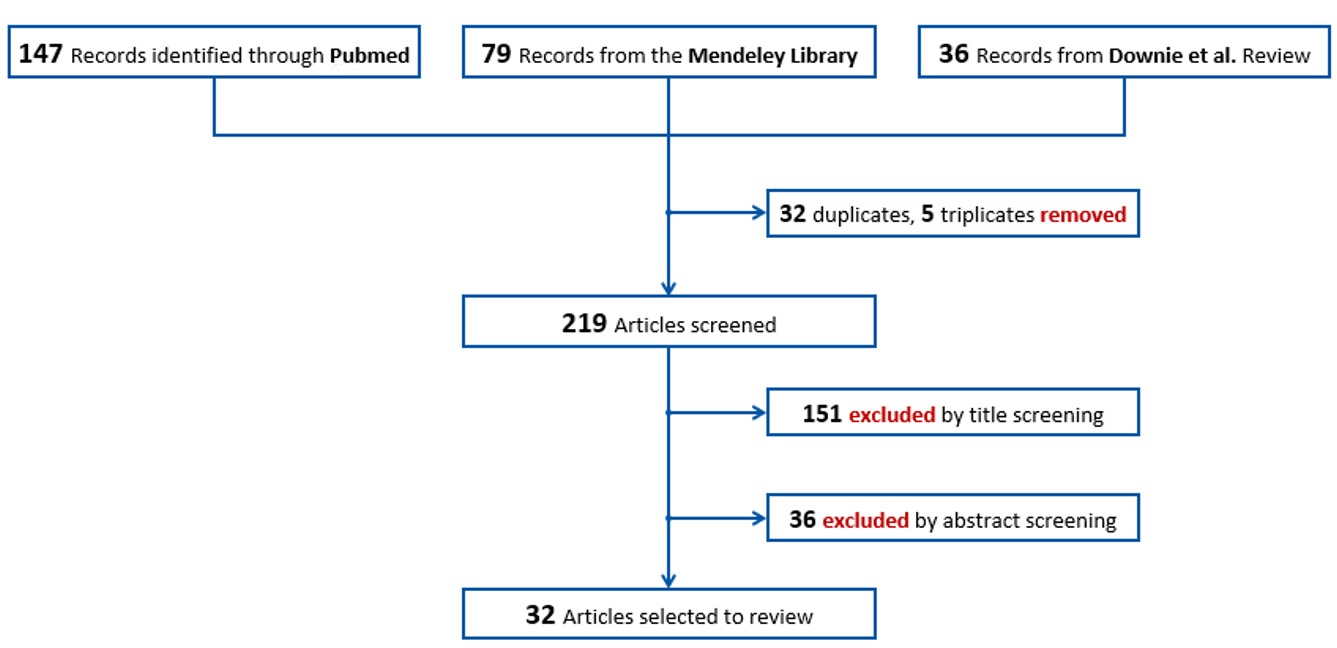
The three sources all together yielded 262 articles, some of which were duplicated in two or all the sources, as shown in Figure 1. The final articles to be screened were 219.
Documents selection
The selection of articles to be included in the review followed two steps, both of which were performed independently by two people:
(1) Titles screening
The first screening was made considering the title of the articles. Articles focusing on one disease only, carrier screening, case reports, protocols only, and infective outbreaks in the Neonatal Intensive Care Unit (NICU) were excluded. After this first step, 151 articles were excluded for relevance reasons.
(2) Abstracts screening
The second step consisted of reading the abstracts and assigning each article to one of 14 pre-identified categories grouped in three areas: clinical, social, and governance [see Figure 2]. Reading the abstracts allowed a stricter selection of articles with a clear focus on the application of Genome Sequencing (GS) to population-wide NBS, while excluding the publications that used GS as a diagnostic tool. Finally, we reduced the redundancy based on the article’s topic and publication date (e.g., for articles on the same topic, the most recent was preferred). After this step, 36 articles were excluded.
The final number of articles retained for the review was 33.
A flowchart of the selection of the articles can be found in Figure 3.
RESULTS
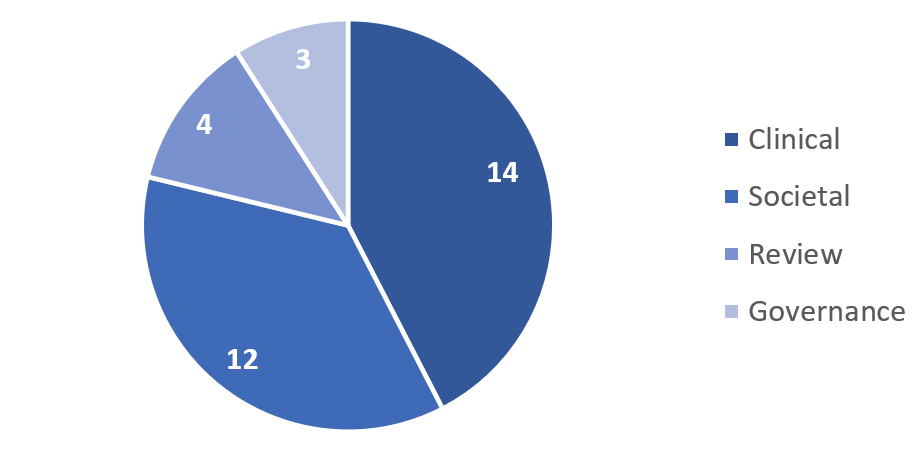
The mixed methods search brought to the identification of 33 articles distributed as in Figure 4.
For the Clinical subject, 14 publications were identified. Five papers were focused on wide GS discussion in the last 15 years[22-24,7]. Methods to manage the genomic data produced in the GS analysis and a definition of the clinical actionable conditions have been explored in three publications[2,6,5]. Results and/or discussions about the impact, feasibility, benefits, and costs of the GS in the clinical care of newborns have been reported in five publications[14,26-29].
For the Social subject, 12 publications were identified. Three records dealt with the BabySeq Project, surveying parents and clinicians involved in the trial, parents who denied participation, and a third one analyzing the changed protocol and the concept of family benefit[8,12,30]. The study NC-NEXUS was also taken into consideration, with a publication regarding a Decision Aid tool to support parents in the decision-making process. If GS is to be implemented in NBS, communication and education are key elements that must be considered and promoted[31]. Opinions from genetics professionals were also considered through a paper that presented a survey to the American College of Medical Genetics and Genomics (ACMG) members[32]. Lastly, public views on the incorporation of GS in NBS were also included[33]. Recommendations by the NSIGH Ethics and Policy Advisory Boards were also a result of the search, sharing their opinion regarding the use of GS applied to diagnostic and universal NBS[34]. Finally, two independent opinions and a parents survey were considered[35-37].
For the Governance subject, three publications were identified[9,38,39]. Two[9,38] have a legal focus, analyzing the constitutional framework for the adoption of GS-based NBS programs in the US. The third paper[39] has a policy perspective and lists eight recommendations for the introduction of GS in NBS. These recommendations were elaborated by the Pediatric Task Team of the Global Alliance for Genomics and Health.
Clinical
Wilson and Jungner originally defined the screening criteria to guide the selection of conditions that would have been suitable for screening. Among these criteria, early-stage detectability and treatment availability are still solidly respected. However, the advent of the genomic era with advanced medical technologies and the increased interest in genome screening requested a revision of Wilson and Jungner screening criteria[7]. Certainly, the screening criteria should be further and constantly discussed to reflect people’s evolving interests and needs.
The clinical utility of genetic testing and the efforts to guarantee transparency and quality of the results have been widely discussed in Europe and the USA. The Public and Professional Policy Committee (PPPC) and the Quality Committee of the European Society of Human Genetics (ESHG) addressed these challenges in the past years, and the final recommendations were approved and published in December 2012[24]. Whole-genome analysis might be applied in several circumstances, such as diagnosis in symptomatic patients, research, pharmacogenomics, investigation in pre-symptomatic patients, and population screening programs. In order to develop best practices in implementing WGS/WES into health care:
(1) Stakeholders from different fields should participate in the discussions about WGS/WES implementations, sharing their experience and contributing to the development of national and international guidelines;
(2) A targeted approach should be adopted to avoid unsolicited findings, e.g., known genetic variants with limited or no clinical utility;
(3) WGS/WES analysis should be applied when necessary, ensuring the balance of benefits and limitations for the patient. Genetic experts should explain the benefits and limitations of genetic testing for screening, informing prospective parents and raising public awareness;
(4) A protocol is essential to guide the communication of secondary findings and report how the data will be shared and stored;
(5) Guidelines for informed consent on genomic testing, sample uses (e.g., research studies) and storage need to be developed and widely shared within the appropriate workforce;
The European initiative EuroGentest was established by the European Commission to promote accurate and high-quality genetic diagnostics across Europe, and it was integrated as a working group with the European Society of Human Genetics (ESHG), with whom in 2016 they published the guidelines for diagnostic applications of Next Generation Sequencing (NGS) for rare genetic diseases, consisting of 38 statements with a particular focus on WES and sequencing on selected genes identifying small germline variants (Single Nucleotide Variants (SNVs) and insertions/deletions). In 2021, an update of EuroGentest guidelines for NGS has been published, including five additional statements (a total of 44 statements) by the Solve-RD, a Horizon2020-funded project, born with the aim of finding a diagnosis for a large number of rare diseases (www.solve-rd.eu)[22].
GS-based NBS pilot projects
The implementation of GS in newborns triggered great interest in the setting of explorative pilot projects to assess medical, economic, ethical, and social impact in the healthcare system and among the general population.
The BabySeq project (ClinicalTrials.gov Identifier: NCT02422511) is a randomized trial on newborns with the aim to assess the impact of genomic sequencing in the newborn period to screen healthy infants for current and future health risks and provides data about the feasibility, risks, benefits, and costs of the integration of exome sequencing in the clinical care of newborns. The BabySeq2 Project (ClinicalTrials.gov Identifier: NCT05161169) is currently in the recruitment phase and aims to expand and improve the results obtained in the first study. Results reported for the BabySeq project were obtained by the clinical trial on 159 children from the well-baby nursery at Brigham and Women’s Hospital (127 healthy newborns) and from the neonatal and pediatric intensive care units at Boston Children’s Hospital in Massachusetts General Hospital (32 ill newborns)[28]. 1,514 genes [Supplementary List 1] were curated and classified into three categories (A, B, or C). Category A includes genes with definitive or strong evidence to cause a highly penetrant childhood-onset disorder; Category B includes genes based on actionability during childhood; Category C includes genes that did not meet criteria to be returned in the newborn genome sequencing report[6]. A table including an example of genes from category A from Ceyhan-Birsoy et al. (2017) has been appended[28] [Supplementary Table 1].
After testing, a newborn genomic sequencing report is generated, including information on pathogenic and likely pathogenic variants, monogenic disease variants, recessive carrier variants for childhood-onset or actionable conditions, and pharmacogenomic variants. The analysis also contains information on variants of uncertain/unknown significance (VUS) indications. However, only a randomized group of families received newborn GS reports and the results obtained from the study were disclosed to the newborn’s parents during an in-person consultation by a genetic counselor and physician. The reports are available in both hospitals and online through a GeneInsight Clinic instance[14].
In the BabySeq project, WES analysis uncovered the risk of childhood-onset diseases in 15/159 (9,4%) of newborns, and none of these was expected based on the clinical histories of babies and their parents. Only parents of 85/159 newborns accepted to receive information on adult-onset actionable conditions, and in 3/85 cases a risk was identified. 88% of newborns were carriers of recessive disease and 5% were carriers of pharmacogenomics variants. Among the newborns with carrier-status variants, 8 of 140 (6%) also had VUS in one of the reported carrier genes. The number of carrier-status variants ranged from one to seven variants in a single newborn[28].
Regarding the yield of the GS approach compared to standard NBS methods, the BabySeq project’s results were discordant compared with conventional NBS and NBS plus WES[29]: 84% of newborns were NBS and WES negative; 1/159 infants were positive for the same disorder by both approaches; 9/159 infants were NBS positive and WES negative. Among the latter, 7 were reported as false positives after subsequent analysis. 15/159 infants were WES positive and NBS negative, indicating the risk of genetic conditions not detectable through the conventional NBS approach[29]. However, the BabySeq project results demonstrated the efficacy of newborn GS in detecting risk and carrier status for a wide range of disorders that cannot be detected by current NBS assays[28].
The North Carolina Newborn Exome Sequencing for Universal Screening (NC NEXUS) project (ClinicalTrials.gov Identifier: NCT02826694) was concluded in 2020 and examined the use of WES for NBS versus the conventional NBS approach. 106 infants were enrolled, including two cohorts: 61 healthy infants whose parents were approached for participation in the study prenatally and 45 ill infants affected by inborn errors of metabolism (17) and hearing loss (28), already detected by conventional NBS methods. Trio analysis was not performed. However, a follow-up parental sequencing has been performed in cases for which compound heterozygosity was suspected.
In the NC NEXUS project, WES correctly identified 88% of the cases with already diagnosed metabolic disorders and only 18% with already diagnosed hearing loss. Moreover, actionable findings that would not have been revealed by conventional NBS were revealed in four newborns. Some parents were selected to receive additional information about childhood-onset conditions with low or no clinical actionability, clinically actionable adult-onset conditions, and carrier status for autosomal-recessive conditions[27]. Carrier findings in newborns whose parents requested this information were detected with an average of 1.8 per infant (with a maximum of 7 variants).
Clinical actionability was detected using the age-based semiquantitative metric[5].
Conditions were categorized into four categories:
(1) Pediatric conditions with high medical actionability;
(2) pediatric conditions with low or no medical actionability;
(3) adult conditions with high medical actionability;
(4) adult conditions with low or no medical actionability.
According to these criteria, 755 gene-disease pairs were categorized (the list of 755 genes from Milko et al. (2019) has been included [Supplementary List 2][5]. An abnormal or positive screen GS-NBS result related to high medical actionability conditions was reported by observing likely pathogenic and/or pathogenic variants in genes associated with pediatric conditions. A normal or negative GS-NBS result was defined by the absence of likely pathogenic or pathogenic variants. Positive results were associated with the presence of likely pathogenic or pathogenic variants found in gene(s) reported in the metabolic or hearing loss diagnostic list. Inconclusive results included, for example, a single heterozygous variant found in a gene associated with an autosomal-recessive condition and/or variants of uncertain significance (VUS) in genes on the diagnostic list. Negative results indicated no detection of any pathogenic or likely pathogenic variants or any VUS on the diagnostic gene lists. 15/17 (88,2%) of patients affected by metabolic conditions resulted as GS-NBS positive. In the hearing loss cohort, “inconclusive” findings, not providing definitive results, were reported (some participants were heterozygous or homozygous for different VUSs in genes associated with hearing loss). Two false negative results were detected: one patient had a single heterozygous pathogenic variant in a gene associated with maple syrup urine disease and a patient with Malonyl-CoA decarboxylase deficiency had a homozygous missense VUS. However, since the authors did not have sufficient information to better identify the genetic etiology of the patient’s disease, both were reported as “inconclusive findings”. One patient was a carrier for another condition. 5/28 (17,9%) patients affected by hearing loss tested GS-NBS positive and two of them had positive screen results unrelated to their condition[27].
After the conclusion of the NC NEXUS project, it has been stated that using a GS approach could not widely substitute current screening tests. However, genomic information could be useful to perform a “secondary” or “indication-based” analysis, improving the sensitivity and specificity of NBS for inborn errors of metabolism[27].
In the Netherlands, the NBS (NGSf4NBS) project is a technical feasibility study also aiming at assessing the ethical, legal, social, and financial aspects to explore the adoption of NGS approaches as a first-tier method in NBS[26]. The study will proceed in three steps. In Step 1, inherited metabolic disorders eligible for NGS as a first-tier test will be identified based on treatability. In Step 2, the feasibility, limitations, and comparability of different technical NGS approaches and analysis workflows for NBS will be tested. In Step 3, the results will be incorporated into the current Dutch NBS program, including guidelines for the referral of a child after a positive NGS test result[26].
Methods to evaluate the criteria for inclusion of genes in GS studies
NBS through WGS and WES should be based on a clear path of clinical utility and/or actionability[23]. The magnitude of the genomic information generated, and its management are key challenges of introducing GS in the clinical setting. Other issues that must be taken into account are the definition of a subset of clinically actionable findings, the use of standardized protocols, and the introduction of appropriate and shared informed consensus for the families involved. In 2016, Berg et al. defined a semiquantitative metric for evaluating clinical actionability by assessing five criteria: the severity and likelihood of manifesting a particular condition, the efficacy and acceptability of the intervention, and the overall knowledge base of the gene-disease association[2]. The metric did not take into account the individual’s age and sex, the timing of the onset of the disease, and the availability and cost of any preventive strategy.
The North Carolina Newborn Exome Sequencing for Universal Screening (NC NEXUS) project implemented the semiquantitative metric and assessed an age-based framework for evaluating genome-scale sequencing results in NBS. The age-based, semiquantitative metric categorized gene-disease pairs into groups based on age of onset or timing of interventions, improving the past method and facilitating the definition of inclusion criteria in the GS studies[5].
Additionally, a list of genes with putative pediatric relevance based on the framework released by the Clinical Genome Resource (ClinGen) working group has been assessed to manage the return of results in the BabySeq project. The generation of the gene-disease pair association was curated for the following criteria: validity of gene-disease association, age of onset, penetrance, and inheritance pattern. Based on the selected criteria, three categories of classification of gene-disease pairs were defined: category A: genes included in the newborn genomic sequencing report with definitive or strong evidence to cause a highly penetrant childhood-onset disorder; category B: genes included in the newborn genomic sequencing report based on actionability during childhood; category C: genes that did not meet criteria to be returned in the newborn genomic sequencing report[6].
A comparison between the NC NEXUS age-based framework and the BabySeq categorization approach revealed differences in the methods used to define each category. The NC NEXUS age-based semiquantitative metric includes several components to achieve actionability score criteria, whereas the BabySeq criteria differ between each of the three categories. BabySeq category A is focused on clear evidence of gene-disease relationship without actionability considerations. Category B includes potential actionability. Category C includes low penetrance, insufficient evidence or late-onset conditions, and non-invasive intervention in childhood. A solution proposed was to report actionable genomic information at the corresponding age-appropriate stage (e.g., infancy, childhood, adult) to overcome any potential social, ethical, or psychological issue related to non-actionability conditions[5].
Societal
Incorporating WGS/WES into population-wide NBS programs triggers significant ethical and policy concerns, as it implies the generation of incidental health information of known and unknown clinical significance for millions of infants annually[36]. When implementing a new technology in a state-run program, it is particularly important to reach clarity in the evaluation of benefits and limitations. This is notably valid when the technology is GS, as test results present a heterogeneous, complex, and unsure nature[33].
Conventional (biochemical-based) NBS is considered a standard of care and is often a mandatory, state-supported activity, e.g., in Canada and the US, where parental consent is typically implied[40,41]. Introducing NGS technologies could dramatically change the context, shifting the balance between clinical benefits and risks and raising new questions that could threaten the universality and moral authority of NBS. GS technology has raised fundamental challenges to the traditional ways genomic information is communicated. If GS was to be incorporated into standard NBS practice, clinicians, public health officials, and other stakeholders would need to agree on the type of information that they should seek and communicate to parents[31].
Ulm et al. in 2015 surveyed members of the American College of Medical Genetics and Genomics (ACMG) to gather genetic professionals’ opinions regarding the use of WGS in NBS[32]. Starting from the premise that 86% of the respondents believe WGS should not be included in NBS yet, many critical challenges were identified, such as the introduction of pre- and post-counseling, the interpretation of results, and follow-up access. Informed consent should be required from parents to enable them to decide which information to receive but with the confidence of knowing that laws and policies are being implemented to protect against discrimination and privacy[32]. It is interesting to notice that at the time the participants filled out the survey (November-December 2012), 28% believed WGS would have been implemented in 5 years (by 2017) and 23% in 6-10 years (by 2018-2022).
Informed consent and return of results
Given the nature of NBS, for which the primary beneficiary is the newborn, parents have a substantial role in the process. Joseph et al. conducted four focus groups with socioeconomically and ethnically diverse pregnant women to examine their views and perspectives regarding the potential application of WGS to NBS. For many women, knowledge and information are fundamental tools to have a sense of control over labor and childbirth - and consultations and education regarding NBS are key topics of conversation that should happen before the test, in order to understand the process and have the opportunity to ask questions[37]. Formal permission or written consent was, however, a secondary priority for parents, while it was felt more urgent in case NBS was performed with WGS, given the increased complexity of genetic information. The need for formal parental permission implies the possibility that parents opt out, thus altering the universality principle that characterizes NBS[34].
Genetti et al. in 2018 evaluated parental interest in a randomized trial of GS-NBS, in particular analyzing causes for declining participation, before and after an enrolment meeting with a genetic counselor. Risk communication was found to be a key element during the education process for informed consent, given the sensitivity of genetic information and the apprehension that this information would be recorded in their infants’ medical documents[30].
Psychological distress
Families and professionals involved in newborn genetic screening are challenged with complex and onerous questions that can lead to an increased amount of new knowledge which can be difficult to deal with. Parents have the authority, both legal and moral, of making decisions for their newborns, including medical decisions that are, supposedly, in their child’s best interest. When using GS, a large number of gene variants are possibly detected, including genes encoding for adult-onset disorders. Such timing of testing, being in the neonatal period, makes it impossible for the primary beneficiaries, i.e., newborns, to make their own decisions depriving them of future adult autonomy and confidentiality[11,12,35,37].
While the use of GS as a diagnostic tool is accepted, the uncertainty and ambiguity of some results of GS as a screening tool could transform healthy newborns into pre-sick or “patient-in-waiting”[42], risking premature medicalization of infants and causing significant distress and worry in parents[34].
Many other potential drawbacks for the screened family are the damage to the child’s self-esteem, stigmatization, and the sense of guilt of transmitting a pathogenic variant to your child; this information could also be the cause of discrimination, lack of privacy in different circumstances, with issues accessing medical insurance being the first difficulties on a potentially long list[35].
Genetic professionals and laboratorians are also suffering from potential moral and ethical dilemmas: Ross et al. in 2019 reported a case in the BabySeq project where the discovery of an actionable adult-onset disease in a newborn led to a dilemma of the personnel that could not return a result that was widely considered actionable[12]. On the basis of this case, the BabySeq protocol was then modified, invoking the principle of family benefit, for which the best interest of the child includes his parents’ well-being. Following these modifications, parents could decide whether they wanted to receive information on adult-onset variants, even though it is still widely accepted[43] that children should not be tested for adult-onset conditions. For Ross & Clayton, one solution could have been to modify the BabySeq analytical process in order not to discover those variants, designing the study to limit the search to relevant genes and reduce the risk of finding stress-inducing information[12].
A survey conducted by Pereira et al. published in 2019 demonstrated that parents and clinicians would prefer NBS without GS, even though parents showed more trust than clinicians towards GS. This shows that what is considered a clinical benefit to the clinicians is different from the perception of the parents (i.e., parental/personal utility), which might have a broader range of expectations, showing once again how relevant and crucial the education process is in these circumstances[8].
Considering how fast today’s society evolves and how complex and sensitive this field is - more frequent societal consultations are key to understanding whether there is a community consensus.
Governance
The psychological distress and worry around using GS in NBS bring governance and policy consequences that must be taken into consideration. For example, parental worry could cause follow-up visits, tests, and services that may not be medically indicated[36].
Moreover, when clinicians or other healthcare professionals have the role of returning results to patients, time management is a concern, since counseling parents and educating them on procedures and next steps will be time- and energy-consuming and, therefore, costly. It has to be taken into consideration that all positive screen results will need follow-up care, confirmatory testing, and monitoring, ensuing even more time and costs to the healthcare system[34].
Genetics professionals surveyed by Ulm et al. think that the complexity implied in the use of GS in NBS should lead to a new counseling paradigm, forcing a non-mandatory program that envisages consent and the option to opt out in a setting where genetic discrimination is prevented[32]. These changes and challenges should thus require a new setting and an infrastructure boosting education and training of the workforce involved[35].
On the same line, two papers[9,38] analyzed the US legal framework with respect to the introduction of GS in NBS programs. Both concluded that the current “constitutional boundaries” do not allow the introduction of mandatory neonatal screening programs using GS. The first argument is that mandatory screening is based on two fundamental legal bases:
(1) Police power that allows the state to intervene in order to protect the health and safety of citizens AND
(2) Parens patrie that allows public authority to make decisions in the best interest of the children despite the opinion of the parents.
Both principles do not seem to be applicable to genomic screening unless it is limited to a strict number of genes (and variants on those genes) that cause severe but treatable conditions with an almost certain pediatric onset[9,38,39].
From a health policy perspective, there is a consensus regarding the introduction of GS-based NBS programs which should not substitute the current conventional NBS programs, meaning that the costs for implementing the new program are on top of the existing one with limited overlap[39]. Another important aspect considered by all the three papers[9,38,39] is equity: despite being subject to consent from the parents, once introduced, GS-based NBS should be equally accessible to all newborns. An interesting concept linked with equity concerns is the possibility for the families to have raw data from GS analyzed and interpreted independently; if families can get access to raw data, some of them, the wealthier and more educated, could look for deeper analysis and interpretation even for a portion of the genome not included in the NBS program. Is that ethical? Is that fair, considering that other families will not have that possibility?[9]
LIMITATIONS
The rapid evolution of the field and the increasing number of pilot programs using GS for NBS make it difficult to give a snapshot without the risk of missing the most recently published evidence. To make an example, while preparing this manuscript, a rapid evidence review on the implementation of large-scale genomic screening was published by Alarcón Garavito et al.[44].
Moreover, the decision to focus exclusively on NBS programs using WES or WGS forces to neglect some works on disease-specific genetic screening that could provide some additional evidence, especially on topics such as acceptability by the parents and management of incidental findings and VUS.
Finally, for this work, only peer-reviewed articles were taken into consideration. This could have limited the identification of relevant information, especially on governance and legal aspects that could have been included in grey literature, such as project public deliverables, reports, and policy guidelines.
CONCLUSIONS
Although there is a broad and animated debate on the use of GS for NBS, there is still little real-world evidence available from a few pilot projects (namely BabySeq and NC-Nexus, both carried out in the USA). Other pilot projects have been recently launched in Europe and the UK and more evidence will become available in the coming years. Despite a consensus in the literature on the key principles that should guide the use of GS in NBS, many important issues are still to be adequately addressed and solved.
All authors agree that NBS should include only actionable genes, but the definition of actionable is still a matter of debate, as well as the criteria and ideal frequency of updates of the list of genes-diseases to be screened for. Currently, informed consent from the parents seems to be the preferred approach, but there is still an open discussion on how to manage incidental findings or information on the status of the carrier.
Ethical, legal, social, and budgetary issues still constitute great challenges and major barriers to the wide, equitable, and uniform adoption of GS in NBS. When looking at these aspects, it is important to also consider the other side of the coin, i.e., the burden that inherently accompanies a family who did not get the chance of an early diagnosis or the management of critically ill patients in NICUs. Early diagnosis could also generate cost savings for the healthcare systems as it allows them to prevent severe symptoms that may require frequent hospitalizations. These savings could at least partially balance the additional costs generated by GS-NBS, which, according to the majority of authors, should not substitute the current NBS programs but run in parallel as additional screening. Unfortunately, it was not possible to find any published studies with information on cost-effectiveness and the estimation of potential savings of healthcare resources by using GS in NBS.
The management of genomic data of newborns for secondary use (e.g., for research purposes) should be balanced with the right of children to an “open future” and to autonomously make decisions on the use of their own genomic profile. As shown by this literature review, no easy or straightforward solutions have emerged so far. Moreover, a one-size-fits-all approach will probably never work, as GS-based NBS should take into consideration the specific value and ethical frame of the community where it is deployed. Ten years ago, 50% of the surveyed experts of the ACMG expected GS to be implemented in the NBS everyday practice. Evidently, we are not there yet. Further pilots and consultations with the stakeholders will be necessary before GS-based NBS programs can be widely implemented.
DECLARATIONS
Acknowledgments
The authors would like to thank Virginie Bros-Facer, David Pearce, and Daniel Scherman for their precious support. Special thanks also to Maria Iascone for the valuable suggestions.
Authors’ contributions
Design of the methodology and the search algorithm: Magnifico G, Benvenuti S
Blind selection of the articles: Magnifico G, Artuso I
Writing of the manuscript: Magnifico G, Artuso I, Benvenuti S
Availability of data and materials
Not applicable.
Financial support and sponsorship
This work was supported in part by Regione Lombardia, ITALY [FT5132], and by Fondazione Telethon [GSP21003].
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2023.
Supplementary Materials
REFERENCES
1. Loeber JG, Platis D, Zetterström RH, et al. Neonatal screening in europe revisited: An ISNS perspective on the current state and developments since 2010. Int J Neonatal Screen 2021;7:15.
2. Berg JS, Foreman AK, O'Daniel JM, et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet Med 2016;18:467-75.
3. Berg JS, Agrawal PB, Bailey DB Jr, et al. Newborn sequencing in genomic medicine and public health. Pediatrics 2017:139.
4. Burlina A, Jones SA, Chakrapani A, et al. A new approach to objectively evaluate inherited metabolic diseases for inclusion on newborn screening programmes. Int J Neonatal Screen 2022;8:25.
5. Milko LV, O'Daniel JM, DeCristo DM, et al. An age-based framework for evaluating genome-scale sequencing results in newborn screening. J Pediatr 2019;209:68-76.
6. Ceyhan-Birsoy O, Machini K, Lebo MS, et al. A curated gene list for reporting results of newborn genomic sequencing. Genet Med 2017;19:809-18.
7. Andermann A, Blancquaert I, Beauchamp S, Déry V. Revisiting Wilson and Jungner in the genomic age: a review of screening criteria over the past 40 years. Bull World Health Organ 2008;86:317-9.
8. Pereira S, Robinson JO, Gutierrez AM, et al. BabySeq Project Group. Perceived benefits, risks, and utility of newborn genomic sequencing in the babyseq project. Pediatrics 2019;143:S6-S13.
9. Zacharias RL, Smith ME, King JS. The legal dimensions of genomic sequencing in newborn screening. Hastings Cent Rep 2018;48 Suppl 2:S39-41.
10. Tarini BA, Goldenberg AJ. Ethical issues with newborn screening in the genomics era. Annu Rev Genomics Hum Genet 2012;13:381-93.
11. Lantos JD. Ethical and psychosocial issues in whole genome sequencing (WGS) for newborns. Pediatrics 2019;143:S1-5.
12. Ross LF, Clayton EW. Ethical issues in newborn sequencing research: the case study of babyseq. Pediatrics 2019:144.
13. Pichini A, Ahmed A, Patch C, et al. Developing a national newborn genomes program: an approach driven by ethics, engagement and co-design. Front Genet 2022;13:866168.
14. Holm IA, Agrawal PB, Ceyhan-Birsoy O, et al. BabySeq project team. the babyseq project: implementing genomic sequencing in newborns. BMC Pediatr 2018;18:225.
15. Schuijff M, Dijkstra AM. Practices of responsible research and innovation: a review. Sci Eng Ethics 2020;26:533-74.
16. Loeber A, Bernstein MJ, Nieminen M. Implementing responsible research and innovation: from new public management to new public governance. In: Blok V, editor. Putting Responsible Research and Innovation into Practice. Cham: Springer International Publishing; 2023. pp. 211-28.
17. Responsible research and innovation: Europe’s ability to respond to societal challenges Available from: https://data.europa.eu/doi/10.2777/11739 [Last accessed on 24 Aug 2023].
18. European commission. Open innovation, open science, open to the world. Available from: http://europa.eu/rapid/press-release_SPEECH-15-5243_en.htm [Last accessed on 24 Aug 2023].
19. HTA core modelR version 3.0. Available from: https://www.eunethta.eu/hta-core-model/ [Last accessed on 29 Aug 2023].
20. HTA core model version 3.0. Available from: https://www.eunethta.eu/wp-content/uploads/2018/03/HTACoreModel3.0-1.pdf [Last accessed on 29 Aug 2023].
21. Downie L, Halliday J, Lewis S, Amor DJ. Principles of genomic newborn screening programs: a systematic review. JAMA Netw Open 2021;4:e2114336.
22. Souche E, Beltran S, Brosens E, et al. Recommendations for whole genome sequencing in diagnostics for rare diseases. Eur J Hum Genet 2022;30:1017-21.
23. Hendricks-Sturrup RM, Lu CY. When should genomic and exome sequencing be implemented in newborns? Genet Med 2020;22:809-10.
24. van El CG, Cornel MC, Borry P, et al; ESHG Public and Professional Policy Committee. Whole-genome sequencing in health care: recommendations of the European society of human genetics. Eur J Hum Genet 2013;21:580-4.
25. Morava E, Baumgartner M, Patterson M, Peters V, Rahman S. Newborn screening: to WES or not to WES, that is the question. J Inherit Metab Dis 2020;43:904-5.
26. Veldman A, Kiewiet MBG, Heiner-Fokkema MR, et al. Towards next-generation sequencing (NGS)-based newborn screening: a technical study to prepare for the challenges ahead. Int J Neonatal Screen 2022;8:17.
27. Roman TS, Crowley SB, Roche MI, et al. Genomic sequencing for newborn screening: results of the NC NEXUS project. Am J Hum Genet 2020;107:596-611.
28. Ceyhan-Birsoy O, Murry JB, Machini K, et al. BabySeq Project Team. Interpretation of genomic sequencing results in healthy and ill newborns: results from the BabySeq project. Am J Hum Genet 2019;104:76-93.
29. Wojcik MH, Zhang T, Ceyhan-Birsoy O, et al. BabySeq Project Team. Discordant results between conventional newborn screening and genomic sequencing in the BabySeq Project. Genet Med 2021;23:1372-5.
30. Genetti CA, Schwartz TS, Robinson JO, et al. BabySeq Project Team. Parental interest in genomic sequencing of newborns: enrollment experience from the BabySeq project. Genet Med 2019;21:622-30.
31. Lewis MA, Paquin RS, Roche MI, et al. Supporting parental decisions about genomic sequencing for newborn screening: the NC NEXUS decision Aid. Pediatrics 2016;137 Suppl 1:S16-23.
32. Ulm E, Feero WG, Dineen R, Charrow J, Wicklund C. Genetics professionals' opinions of whole-genome sequencing in the newborn period. J Genet Couns 2015;24:452-63.
33. Bombard Y, Miller FA, Hayeems RZ, et al. Public views on participating in newborn screening using genome sequencing. Eur J Hum Genet 2014;22:1248-54.
34. Johnston J, Lantos JD, Goldenberg A, Chen F, Parens E, Koenig BA. members of the NSIGHT Ethics and Policy Advisory Board. Sequencing newborns: a call for nuanced use of genomic technologies. Hastings Cent Rep 2018;48 Suppl 2:S2-6.
35. Reinstein E. Challenges of using next generation sequencing in newborn screening. Genet Res 2015;97:e21.
36. Grob R, Roberts S, Timmermans S. Families' experiences with newborn screening: a critical source of evidence. Hastings Cent Rep 2018;48 Suppl 2:S29-31.
37. Joseph G, Chen F, Harris-Wai J, Puck JM, Young C, Koenig BA. Parental views on expanded newborn screening using whole-genome sequencing. Pediatrics 2016;137 Suppl 1:S36-46.
39. Friedman JM, Cornel MC, Goldenberg AJ, Lister KJ, Sénécal K, Vears DF. Global Alliance for Genomics and Health Regulatory and Ethics Working Group Paediatric Task Team. Genomic newborn screening: public health policy considerations and recommendations. BMC Med Genomics 2017;10:9.
40. Morrison A, Dowler J. Newborn screening for disorders and abnormalities in Canada. Available from: https://www.cadth.ca/sites/default/files/pdf/Newborn_Screening_es-26_e.pdf [Last accessed on 24 Aug 2023].
41. Goldenberg AJ, Sharp RR. The ethical hazards and programmatic challenges of genomic newborn screening. JAMA 2012;307:461-2.
42. Timmermans S, Buchbinder M. Patients-in-waiting: living between sickness and health in the genomics era. J Health Soc Behav 2010;51:408-23.
43. COMMITTEE ON BIOETHICS, COMMITTEE ON GENETICS, AND, AMERICAN COLLEGE OF MEDICAL GENETICS AND, GENOMICS SOCIAL, ETHICAL, LEGAL ISSUES COMMITTEE. Ethical and policy issues in genetic testing and screening of children. Pediatrics 2013;131:620-2.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Magnifico G, Artuso I, Benvenuti S. A systematic review of real-world applications of genome sequencing for newborn screening. Rare Dis Orphan Drugs J 2023;2:16. http://dx.doi.org/10.20517/rdodj.2023.17
AMA Style
Magnifico G, Artuso I, Benvenuti S. A systematic review of real-world applications of genome sequencing for newborn screening. Rare Disease and Orphan Drugs Journal. 2023; 2(3): 16. http://dx.doi.org/10.20517/rdodj.2023.17
Chicago/Turabian Style
Magnifico, Giuditta, Irene Artuso, Stefano Benvenuti. 2023. "A systematic review of real-world applications of genome sequencing for newborn screening" Rare Disease and Orphan Drugs Journal. 2, no.3: 16. http://dx.doi.org/10.20517/rdodj.2023.17
ACS Style
Magnifico, G.; Artuso I.; Benvenuti S. A systematic review of real-world applications of genome sequencing for newborn screening. Rare. Dis. Orphan. Drugs. J. 2023, 2, 16. http://dx.doi.org/10.20517/rdodj.2023.17
About This Article
Special Issue
Copyright
Data & Comments
Data

 Cite This Article 6 clicks
Cite This Article 6 clicks













Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.